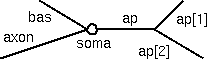

This model will have a soma, an apical dendritic tree with a primary trunk and two distal branches, a basilar tree that is lumped into a single equivalent cylinder, and an axon (see table for dimensions). The soma and axon will have HH spike currents at normal density. The apical dendrites will also have spike currents, but only at 10% of the density in the soma and axon. The basilar dendrites are to be passive.
Geometry Section L
(um)diam
(um)Biophysics soma 20 20 hh ap[0] 400 2 reduced hh* ap[1] 300 1 reduced hh* ap[2] 500 1 reduced hh* bas 200 3 pas axon 800 1 hh *--In the apical dendrites, gnabar_hh and gkbar_hh are reduced to 10% of normal,
and el_hh (equilibrium potential of the hh leak current) is changed to -64 mV.
In the basilar dendrite, e_pas is -65 mV. With these values, resting potential is
approximately -65 mV throughout the cell.
Other parameters: cm = 1 uf/cm2, Ra = 160 ohm cm.
We will follow this sequence of steps.
Step 1. Bring up a CellBuilder
Step 4. Specify geometry
Set up a strategy for specifying geometry
Execute the strategy
Step 5. Specify biophysics
Set up a strategy for specifying biophysics
Execute the strategy
Step 6. Use the model specification
Go back to the main page ("Using the Cell Builder") to work on a different tutorial.
Copyright © 1999-2006 by N.T. Carnevale and M.L. Hines, All Rights Reserved.