
An important aside: when planning a strategy, keep the sequence of subsets and sections in mind. If the order isn't right, or if you need more subsets, then go back to the Subsets page and make the necessary changes.
We begin by dealing with the spatial grid (i.e. discretization or compartmentalization). This is really just a computational issue, not a biological one--we should be focussing on anatomically- and physiologically-relevant subdivisions of the cell, not "how small should the pieces be to get numerical accuracy and stability."
The CellBuilder offers three different ways to make this as painless as possible.
Let's apply the d_lambda strategy to every section in the model. Click on the "all" subset to make sure it is selected, and glance at the shape plot just to check--every section should be red. Then click on the d_lambda checkbox.
Each section of our model is a different size, so we need to specify L and diam individually for every one of them. With versions of NEURON prior to 5.7, this would have required us to select each section individually, and click on its L and diam buttons. That's a lot of clicks.
However, starting with NEURON 5.7 we can save a few clicks by using the "Distinct values over subset" buttons. These tell the CellBuilder that each member of a subset has its own value of L (and/or diam).
So let's stick with the "all" set, and click on the "Distinct values over subset" L and diam buttons.
And if we had a set whose members should have identical lengths, we'd click on the "Constant value over subset" L button, etc..
This is what the strategy should look like now.
Having set up the strategy, we're ready to implement it. But first, save a session file!
The right panel of the Geometry page now contains a set of buttons with numeric fields and spinners which we can use to enter the values of d_lambda, L, and diam. The sequence of these controls parallels their sequence in the middle panel of the CellBuilder.
The first parameter is d_lambda, which controls the spatial grid. Its default value is 0.1, i.e. one tenth of a length constant at 100 Hz, This is short enough for most models, so we can leave it alone. We can always come back later and try a different value if we like.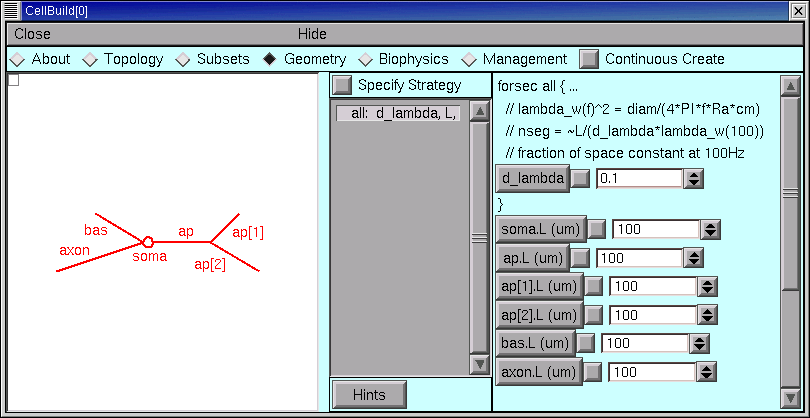
All of the lengths need to be changed.
But where are the diameters?
The drag bar at the right edge is our clue--we only have to pull it down . . .
. . . to see that some diameters also need changing.
Here they are with the desired values.
Time once again to save a session file!
The Biophysics page is next.
Copyright © 1999-2006 by N.T. Carnevale and M.L. Hines, All Rights Reserved.