
For the all subset: click on Ra, cm, and pas
For the apicals subset: click on hh
Note: the apicals already have pas
because the all subset specification will be executed first.
For the axon subset and soma section: click on hh.
Also insert pas because we will need to set g_pas = 0 in the axon and soma
(remember that the all subset inserted pas with a nonzero g_pas).
This crude hack achieves expedience at the cost of clarity--a poor trade at best.
It would have been preferable to define subsets that would make this unnecessary,
i.e. instead of inserting pas into all sections,
do this with a subset called "haspas" that contains all sections except the axon and soma.
"This is left as an exercise for the reader."
Toggle Specify Strategy OFF, and we are ready to enter the desired parameter values.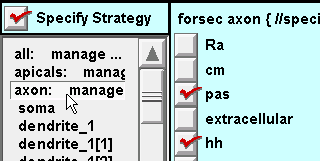
For the apicals subset: set desired hh parameters.
For the axon subset:
default hh is fine.
Execution of the all subset specification inserted pas,
so we must set g_pas = 0 for the axon subset.
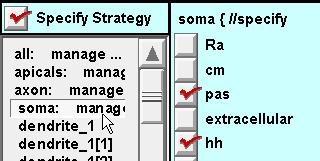
For the soma section: same as for the axon subset.
The model specification is complete. Let's save it to a new session file called pyrfin.ses .
We're ready to test the model cell.
Copyright © 1999-2006 by N.T. Carnevale and M.L. Hines, All Rights Reserved.