First we bring up an Import3d tool, then specify the file that we want to read, take a quick look at what we got, and finally export the data as a NEURON model.
A. We need an Import3D tool.
Open an Import3D tool by clicking on Tools / Miscellaneous / Import3D in the NEURON Main Menu.
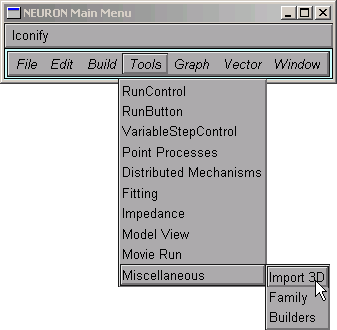
B. We need to choose a file to read.
In the Import3D window, click on the "choose a file" checkbox.
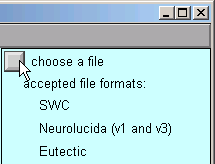
This brings up a file browser. Use the file browser to go to the directory that contains the morphometric data file.
Click on the file's name . . .
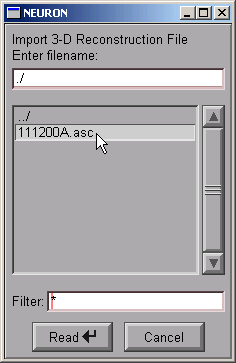
. . . and then click Read.
NEURON's xterm (interpreter window) prints a running tally of how many lines have been read from the data file.
oc> 21549 lines read
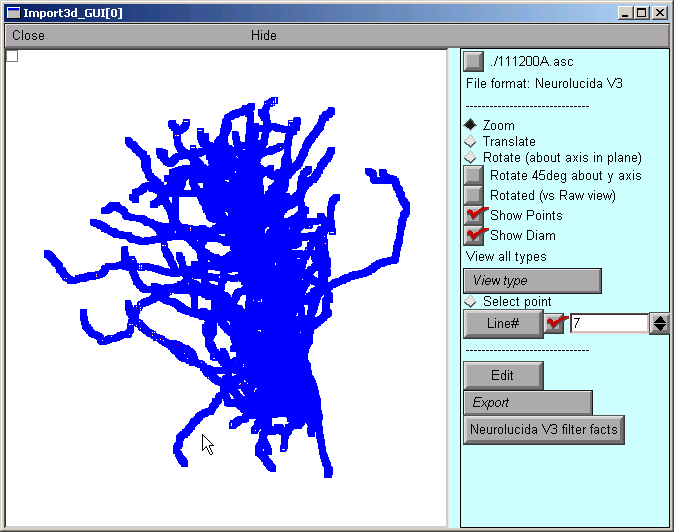
After a delay that depends on the size of the file and the speed of the computer, a figure will appear on the Import3D tool's canvas. Each point at which a measurement was made is marked by a blue square.
If the Import3D tool finds errors in the data file, a message may be printed in the xterm, and/or a message box may appear on the screen. For this particular example there were no errors--that's always a good sign!
The top of the right panel of the Import3D tool will show the name and data format of the file that was read. The other widgets in this panel, which are described elsewhere, can be used to examine and edit the morphometric data, and export them to the CellBuilder or the hoc interpreter.
C. Let's see what it looks like.
It's always a good idea to look at the results of any anatomical data conversion--but those blue squares are in the way!
To get rid of the blue squares that are hiding the branched architecture, click on the Show Points button in the right panel of the Import3D tool. The check mark disappears, and so do the blue squares.
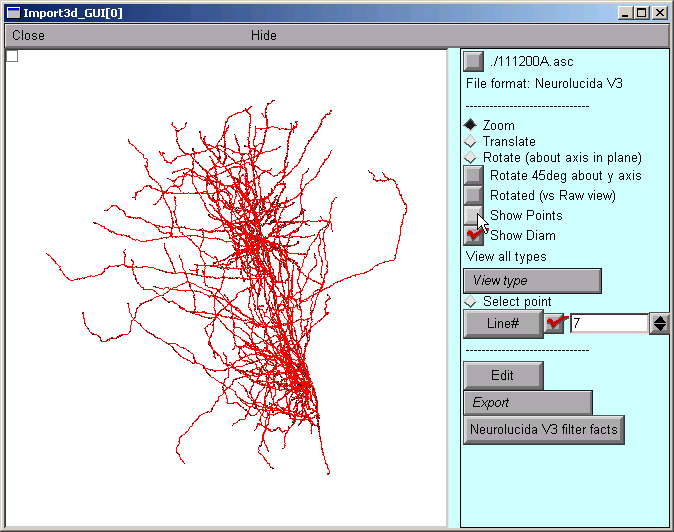
That's a very dense and complex branching pattern.
D. Exporting the model.
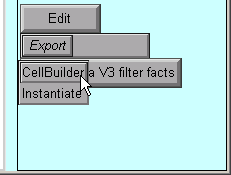
The Import3D tool allows us to export the topology (branched architecture) and geometry (anatomical dimensions) of these data to a CellBuilder, or straight to the hoc interpreter. It's generally best to send the data to the CellBuilder, which we can then save to a session file for future re-use. The CellBuilder, which has its own tutorial, is a very convenient tool for managing the biophysical properties and spatial discretization of anatomically complex cell models.
So click on the Export button and select the CellBuilder option.
But this example has a surprise for us: instead of one CellBuilder, we get two! Under MSWin, they are offset diagonally as shown here, but under UNIX/Linux they may lie right on top of each other so you'll have to drag the top one aside.
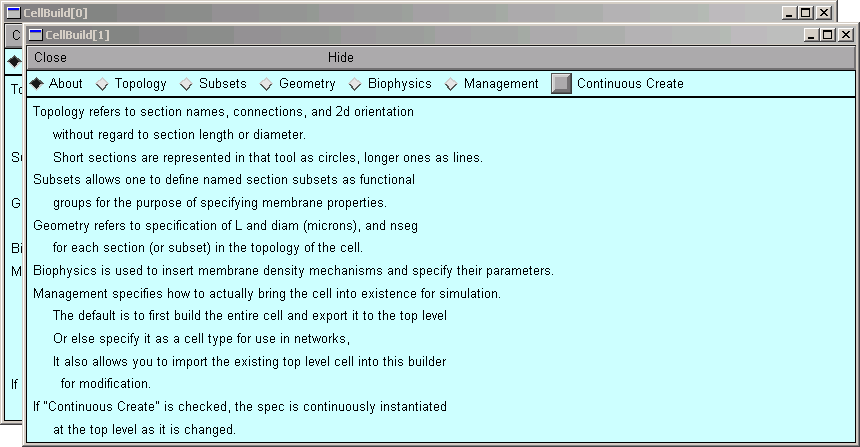
Does getting two CellBuilders mean that the morphometric data file contained measurements from two cells? Maybe that's why the branching pattern was so dense and complex.
But there is an unpleasant alternative: maybe all this data really is from one cell. If there was a mistake in data entry, so that the proximal end of one branch wasn't connected to its parent. one CellBuilder would contain the orphan branch and its children, and the other CellBuilder would contain the rest of the cell.
How can we decide which of these two possibilities is correct?
Examining the Topology pages of these CellBuilders shows us that CellBuild[0] got most of the branches in the bottom half of the Import3D's canvas, and CellBuild[1] got most of the branches in the top half. The morphologies are ugly enough to be two individual cells; at least, neither of them is obviously an orphan dendritic or axonal tree.
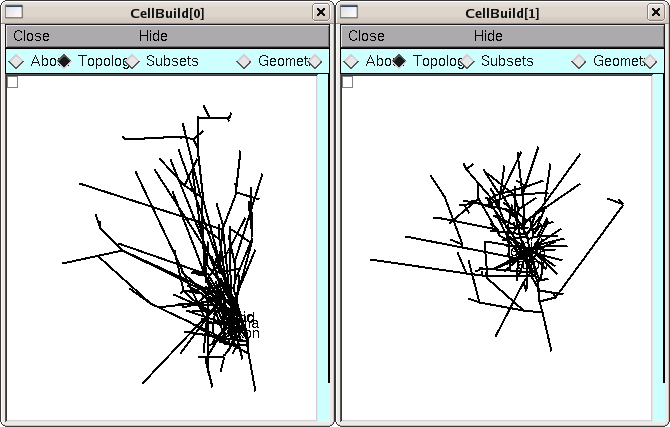
Until we know for sure, it is safest to use the Print & File Window Manager (PFWM) to save each CellBuilder to its own session file. I optimistically called them bottomcell.ses and topcell.ses, respectively.
At this point, we should really use the Import3D tool to closely examine these data, and try to decide how many cells are present. To learn how, work through "Zooming in and out" (one of the "Exploring morphometric data and fixing problems" lessons).