If the CellBuilder's Continuous Create button is checked, changes are passed from the CellBuilder to the interpreter as they occur. This means that you can immediately explore the effects of anything you do with the CellBuilder.
To see how this works, try the following.
1. Start NEURON with its standard GUI in the exercises/cellbuilder directory ( remember how? ).
2. Bring up the CellBuilder and construct a cell that looks like this:
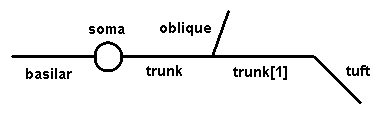
Use any anatomical and biophysical properties you like; these might be interesting to start with:
| Section | L (um) | diam (um) |
|---|---|---|
| soma | 30 | 30 |
| trunk | 400 | 3 |
| trunk[1] | 400 | 2 |
| oblique | 300 | 1.5 |
| tuft | 300 | 1 |
| basilar | 300 | 3 |
- nseg = 1 for all sections
- Ra = 160, Cm = 1, both uniform throughout the cell
- soma has hh
- trunk and all its tributaries have hh, but with gnabar & gkbar reduced by a factor of 10 and gleak = 0 (Subsets makes this easy)
- all dendrites have pas with gpas = 3e-5
3. Toggle Continuous Create ON.
4. In the interpreter verify the structure and parameters of your model with h.topology() and for sec in h.allsec(): print(sec.psection())
5. At the proximal end of tuft, place an alpha function synapse that has onset = 0 ms, tau = 1 ms, gmax = 0.01 umho, and e = 0 mV (hint: NEURON Main Menu / Tools / Point Processes / Managers / Point Manager).
6. Open a graph window to plot soma Vm vs. time. Also set up a space plot that shows Vm along the length of the cell from the distal end of the basilar to the distal end of the tuft.
7. Run a simulation. If necessary, increase Tstop until you can see the full time course of the cell's response to synaptic input.
8. Increase nseg until the spatial profile of Vm is smooth enough (a couple of applications of
for sec in h.allsec():
sec.nseg *= 3in the interpreter window should do the trick). You may need to adjust the peak synaptic conductance in order to trigger a spike. Then use the command
for sec in h.allsec():
print('%s: %d' % (sec, sec.nseg))to see how many segments are in each section.
9. You can also set nseg for any or all sections using the CellBuilder according to options that you select by Geometry/Specify Strategy. You can set the number of segments manually, or let the CellBuilder adjust them automatically according to one of these criteria:
- d_lambda (the maximum length of any segment, expressed as a fraction of the AC length constant at 100 Hz for a cylindrical cable with identical diameter, Ra, and cm)
or - d_X (the maximum anatomical length of any segment)
Try each of these criteria, setting different values for d_lambda or d_X, and see what happens to nseg in each section and how this affects the spatial profile of membrane potential.
- Don't forget that that execution of your strategy is sequential. In other words, if your strategy specifies d_lambda for the all subset, but then sets nseg = 1 for the tuft section, the tuft will end up with nseg = 1 even though it needs a much finer grid according to the d_lambda criterion.
- Whether you choose d_lambda or d_X, the final value of nseg will be an odd number (this preserves the node at x = 0.5).
- Of these two options, d_lambda has a more rational basis is generally preferable.
d_lambda<=0.1is usually adequate.
10. What happens if the sodium channels are blocked throughout the apical dendrites? Use the CellBuilder to reduce apical gnabar to 0 and then run a simulation.
NEURON hands-on course
Copyright © 1998-2018 by N.T. Carnevale and M.L. Hines, all rights reserved.