Physical System
Giant axon from the squid: Loligo pealei
Conceptual Model
Hodgkin-Huxley cable equations
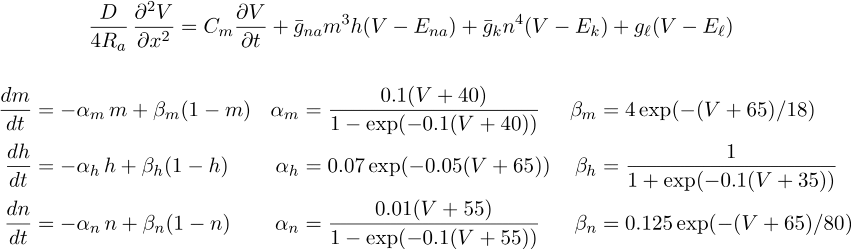
Simulation
Computational implementation of the conceptual model
We could implement this model in Python:
from neuron import h, gui
axon = h.Section(name='axon')
axon.L = 2e4
axon.diam = 100
axon.nseg = 43
axon.insert('hh')But for this exercise, let's instead use the CellBuilder tool to create the model:
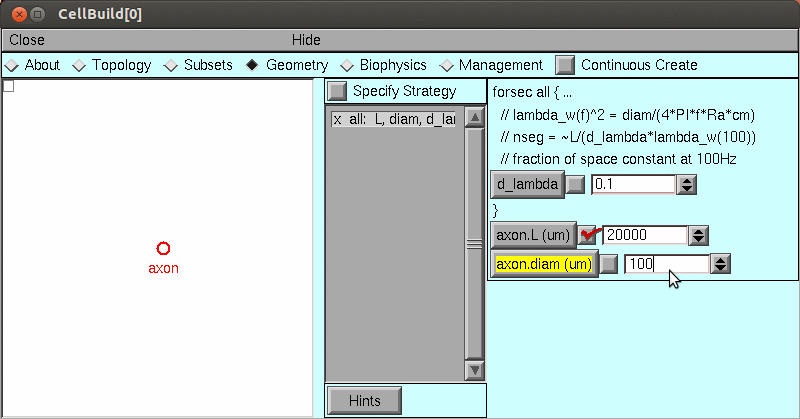
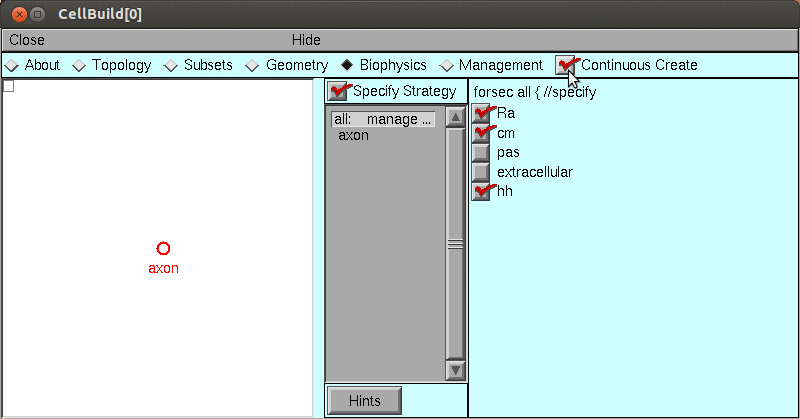
Save the model in hhaxon.ses using NEURONMainMenu / File / savesession.
Using the computational model
If starting from a fresh launch of python, you can load the saved ses file by loading NEURON and its GUI: from neuron import h, gui and then selecting NEURONMainMenu / File / loadsession.
Alternatively you can use NEURON to execute hhaxon.ses
1. change to the appropriate directory in your terminal
2. Start python, and at the >>> prompt enter the commands
from neuron import h, gui
h.load_file('hhaxon.ses')
Exercises
1) Stimulate with current pulse and see a propagated action potential.
The basic tools you'll need from the NEURON Main Menu :
Tools / Point Processes / Manager / Point Manager to specify stimulation
Graph / Voltage axis and Graph / Shape plot to create graphs of v vs t and v vs x.
Tools / RunControl to run the simulation
Tools / Movie Run to see a smooth evolution of the space plot in time.
2) Change excitability by adjusting sodium channel density.
Tool needed:
Tools / Distributed Mechanisms / Viewers / Shape Name
3) Use two current electrodes to stimulate both ends at the same time.
4) Up to this point, the model has used a very fine spatial grid calculated from the Cell Builder's d_lambda rule.
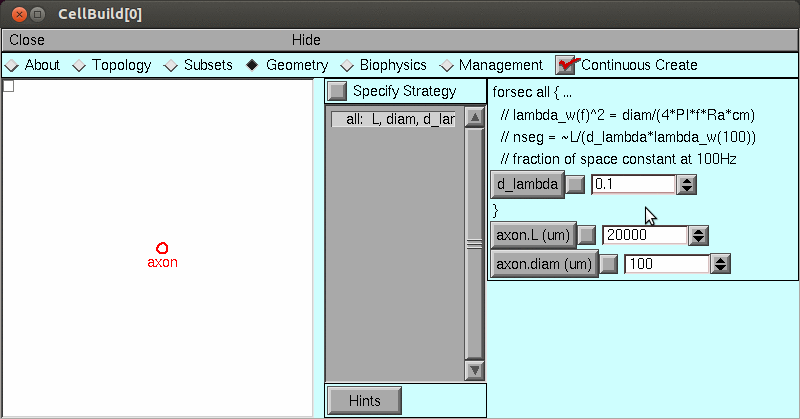
Change nseg to 15 and see what happens.
NEURON hands-on course
Copyright © 1998-2019 by N.T. Carnevale, M.L. Hines, and R.A. McDougal all rights reserved.